PlayMolecule Installation#
PlayMolecule currently only supports Linux Operating Systems (we can provide best support for Ubuntu / CentOS). PlayMolecule sets up a backend for calculations as well as a webserver which can be used to interact with the backend through a user-friendly GUI.
Depending on the installation type these are the minimum system requirements for the server machine. A single-server installation means that calculations will run locally on the server with whatever resources are provided. If molecular dynamics applications need to be run, the server will require at least 1 Nvidia GPU. The more GPUs the more simulations will be able to run in parallel. CPUs can be either Intel or AMD.
On cluster installations the calculations will run on the cluster nodes and thus the server doesn’t require the same computing power and can do without a GPU and fewer cores and RAM. PlayMolecule currently only supports SLURM and SGE clusters. If you require support for a different cluster type please contact info@acellera.com
Installation type |
CPU |
RAM |
HDD |
GPU |
|---|---|---|---|---|
Single Server |
8 Cores |
16 GB |
>500 GB |
Yes/No |
Cluster |
2 Cores |
8 GB |
>500 GB |
No |
Installing PlayMolecule requires an internet connection on the machine so that it’s able to download all the app and service images from the Google Cloud. This is only necessary during the installation and updates of PlayMolecule and not necessarilly during operation (although some apps might require an internet connection for more advanced features like querying the Protein Data Bank). If it’s not possible to provision an internet connection for that machine during the installation please contact us at info@acellera.com and we will provide a zip file with all the required images.
Installing Apptainer#
To install PlayMolecule you will first need to install Apptainer (formerly called Singularity). For installation instructions of Apptainer follow these links:
Make sure you also install the setuid package as per the instructions.
Also ensure that the cryptsetup package is installed on the machine with the following commands
1dpkg -l | grep cryptsetup # For Debian/Ubuntu
2rpm -qa | grep cryptsetup # For EPEL/FEDORA
Obtaining the MAC address#
Before proceeding with the installation of PlayMolecule please send the MAC address of the server
to Acellera so that we can generate a license for your server. You can obtain the MAC address on most
Linux machines by running the ifconfig command. If the command gives multiple MAC addresses please send them
all to Acellera to ensure the license will work correctly.
Creating a service account (Optional)#
It’s not necessary but it’s recommended to create a special Linux user for both the installation and running PlayMolecule.
The software will be installed in the $HOME directory of that account and all the jobs will be submitted from that user to the queueing system.
Here we will assume that the service account user is called svc-playmolecule. Please consult your IT department for creating such a user and
grant them access to your cluster (if applicable).
Installing Miniconda (Optional)#
PlayMolecule installation and management is done through a python program called pmwsdeploy (described in the next section).
This program can be installed in your Operating system’s python installation (assuming it has python 3.7 or above) but ideally we
install it inside a Miniconda environment.
You can find the latest Linux Miniconda installer here.
Download Miniconda under the service account created in the above section (i.e. svc-playmolecule) and then install it by
executing the downloaded .sh file with bash Miniconda3-latest-Linux-x86_64.sh.
Once Miniconda is installed, log out and log in again with the user to activate the Miniconda base environment. If it’s active
you should see (base) on the left side of the console.
Installing PlayMolecule#
Now we can proceed with installing the pmwsdeploy package, which serves to manage our PlayMolecule installation.
First install python if you haven’t already with the following command:
1conda install python
Install it under the svc-playmolecule account with the following command.
Replace [ACELLERA_URL] with the URL provided to you by Acellera.
1pip install acellera-pmwsdeploy --extra-index-url [ACELLERA_URL]
In the email sent to you by Acellera you will find attached a playmolecule-configs.tar.gz.gpg file
as well as a passphrase for decrypting that pgp file. Please download the gpg file and store it in the $HOME
of the svc-playmolecule user.
Now run
1pmwsdeploy
First it will try to detect your Apptainer installation. Once you specify the path to Apptainer it will
ask for the gpg configuration zip which we downloaded above and the passphrase to decrypt it.
After decrypting the configuration file you will face an installation wizard similar to the following:
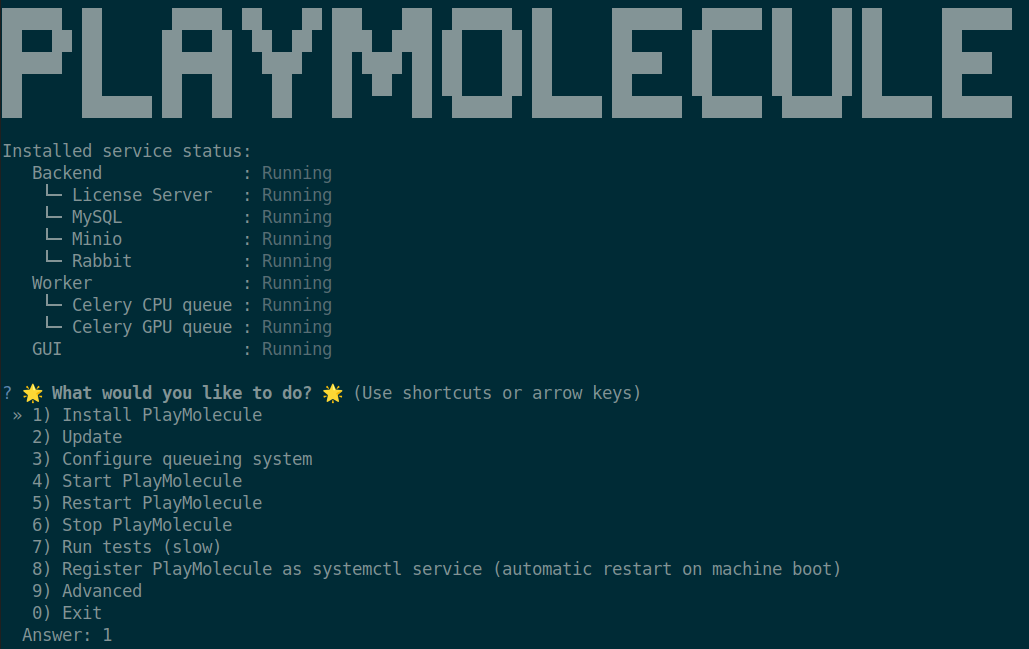
Here select option 1) Install PlayMolecule to proceed with the installation.
For most installations pick the defaults (i.e. install all services on the local machine).
Even if you are installing PlayMolecule with cluster support it’s a good idea to first install it to use Celery
queueing system so that you can test it on the local machine before attempting to configure the cluster connection.
Once all applications and services are installed you can proceed to option 7) Run tests and run a test for ProteinPrepare app
to ensure everything is running as intended. If you get an error please contact info@acellera.com for support.
Ports#
For users outside of the server to access the graphical user interface you will have to open port 7000 on the server.
Then they will be able to access it via the computer’s public IP address (say 192.168.1.19) through their web-browser
via the URL 192.168.1.19:7000.
For users to connect to the PlayMolecule backend via the python API, as well as when installing PlayMolecule for a cluster
you will have to open port 8095 on the server.
Cluster configuration#
Once you have made sure that PlayMolecule runs with the Celery queue, you can proceed to configure the communication settings
with your cluster. For that start pmwsdeploy and head to option 3) Configure queueing system and follow the prompts.
If you are sourcing specific environment files or have set environment variables you will have to provide those. The pmwsdeploy
executable will continously test the cluster connection until the tests pass.
Contact Us#
For any information missing or support needed don’t hesitate to contact us at info@acellera.com
